in vivo crystallization of proteins mediating virulence and/or antibiotic resistance in infectious diseases
Project Leaders:
Christian Betzel, Martin Aepfelbacher, Nicole Fischer, Markus Perbandt and Matthias Wilmanns
Image proof:
Illustration: hegasy.de | © Infectophysics 2019
Scientists of the Infectophysics research group 7 will utilize the potential of living cells, particularly the baculovirus Sf9 insect cell system, to produce from proteins relevant in infection biology bulk amounts of micro- and nanosized protein-crystals.
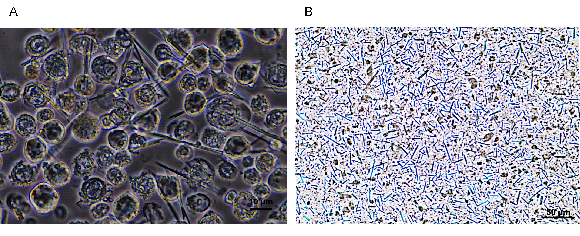
Figure 1. In vivo grown crystals of PAK4:iBox in Sf9 insect cells. (A) Light microscope image of in vivo crystals growing inside living Sf9 cells after 5-6 days post infection. (B) Suspension of purified in vivo grown crystals, ready for diffraction data collection.
Suitable crystals will be applied for the novel method of serial diffraction data collection established recently at PETRA III (DESY) and the European XFEL. Diffraction data obtained will be used to analyse the three-dimensional structure of proteins at high resolution and will deliver crucial information to support drug discovery investigations to target for example bacterial antibiotic resistance (Figure 2).
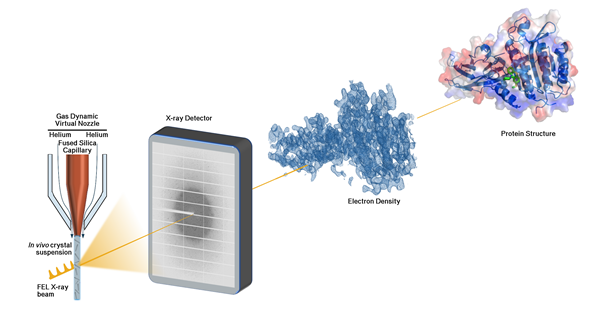
Figure 2. Scheme for serial diffraction data collection and structure based drug discovery investigations.
Till now conventional crystallography requires in principle only one µm sized crystal to collect diffraction data. However, the crystal needs to be shock frozen at 100 K to reduce radiation damage as possible. SFX instead requires approx. 10-9, micro-crystals, which is challenging to produce in vitro, but opens opportunities to avoid radiation damage and to perform time resolved experiments at room temperature to follow for example in 3D enzymes in action.
The Infecto research collaboration 7 will aim to produce X-ray suitable crystals in vivo in high amounts for time resolved studies and will team up with research group 3 to combine efforts to analyse structure and dynamics of infecto relevant proteins at high resolution.
In this context, one major goal of this project will be to obtain more and detailed insights in and about the in vivo crystallization process in cells to establish a platform allowing first to identify promising proteins timely and second allowing to upscale routinely the production of in vivo grown crystals.
Project specific references:
Baskaran, Y. et al. An in cellulo-derived structure of PAK4 in complex with its inhibitor Inka1.
Nature communications 6, 8681, doi:10.1038/ncomms9681 (2015).
Gati, C. et al. Serial crystallography on in vivo grown microcrystals using synchrotron
radiation. IUCrJ 1, 87-94, doi:10.1107/s2052252513033939 (2014).
Jakobi, A. J. et al. In cellulo serial crystallography of alcohol oxidase crystals inside yeast
cells. IUCrJ 3, 88-95, doi:10.1107/s2052252515022927 (2016).
Redecke, L. et al. Natively inhibited Trypanosoma brucei cathepsin B structure determined
by using an X-ray laser. Science (New York, N.Y.) 339, 227-230, doi:10.1126/science.1229663 (2013).
Schönherr, R. et al. Real-time investigation of dynamic protein crystallization in living cells.
Structural dynamics (Melville, N.Y.) 2, 041712, doi:10.1063/1.4921591 (2015).
Tsutsui, H. et al. A diffraction-quality protein crystal processed as an autophagic cargo.
Molecular cell 58, 186-193, doi:10.1016/j.molcel.2015.02.007 (2015).